|
I am a third year CS PhD student Zarlab, UCLA. Prior to this, I spent 4 wonderful years at the Indian Institute of Technology, Delhi (IIT Delhi) studying Biochemical Engineering and Biotechnology and Computer Science. During my undergrad, I was fortunate to have worked with Professor Frank Doyle (Harvard SEAS), Professor Eleazar Eskin (UCLA), and Professor Serghei Mangul (USC). I'm really excited about the making black box ML algos interpretable in the healthcare and genomics space! Seeing algos I've worked on help identify high risk patients, is something that really motivates me. Email / CV / Google Scholar / LinkedIn |

|
|
|
I'm interested in the interdisciplinary areas of Computer Science, Biology and Statistics. I have worked on projects based on Computational Biology, Robotics, Machine Learning, Control Sytstems and Systems Biology in the past. |
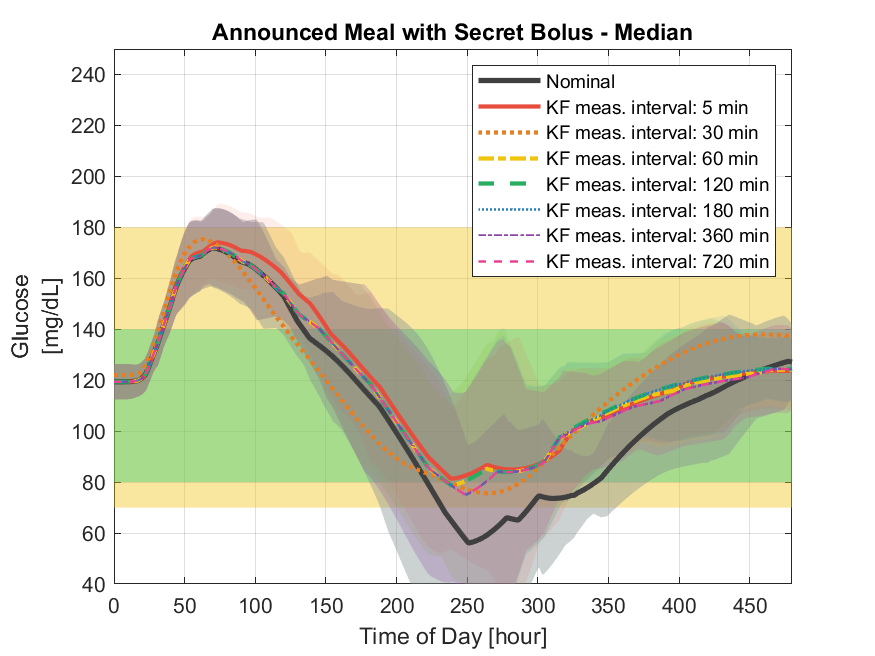 |
Varuni Sarwal, Kelilah L. Wolkowicz, Sunil Deshpande, Joseph Wang, Jordan E. Pinsker, Lori M. Laffel, Mary-Elizabeth Patti, Francis J. Doyle III, Eyal Dassau ATTD, 2019 Identified optimal insulin measurement intervals for insulin sensor design by implementing a feedback-based threshold suspend safety-layer to supplement a zone model predictive control algorithm |
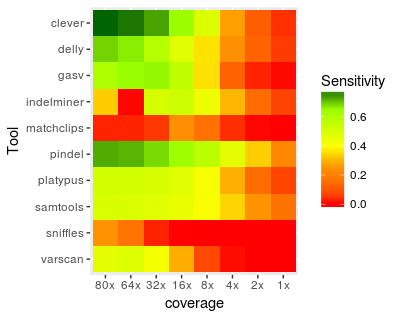 |
Varuni Sarwal, Sei Chang, Ram Ayyala, Sebastian Niehus, Russell Littman, Rahul Chikka, Margaret G. Distler, Eleazar Eskin, Serghei Mangul*, Jonathan Flint* BIG Summer, 2018 Compared the performance of structural variant detection tools against a PCR verified set of deletions to determine tools with the best balance of sensitivity and precision |
|
|
Serghei Mangul, Thiago Mosqueiro, Richard J Abdill, Dat Duong, Keith Mitchell, Varuni Sarwal, Brian Hill, Jaqueline Brito, Russell Jared Littman, Benjamin Statz, Angela Ka-Mei Lam, Gargi Dayama, Laura Grieneisen, Lana S Martin, Jonathan Flint, Eleazar Eskin, Ran Blekhman PLOS Biology, 2019 Estimated the archival stability of computational biology software tools by performing an empirical analysis of the internet presence for 36,702 omics software resources published from 2005 to 2017 |
|
|
|
Hit Counter
|